Today’s lecture brought to you by

Ranae Dietzel & Andee Kaplan

Like a dataframe, but does less
When you print a tibble, you see
width = Inf.
## # A tibble: 5 × 5
## property five_cm ten_cm fifteen_cm twenty_cm
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 pH 7 6.7 6.5 6.4
## 2 NO3 35 22.0 20.0 18.0
## 3 clay 20 20.0 20.0 20.0
## 4 silt 20 20.0 20.0 20.0
## 5 sand 60 60.0 60.0 60.0Plot the change in pH across depth?
gatherWhen column names are values instead of variables, we need to gather
## # A tibble: 5 × 5
## property five_cm ten_cm fifteen_cm twenty_cm
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 pH 7 6.7 6.5 6.4
## 2 NO3 35 22.0 20.0 18.0
## 3 clay 20 20.0 20.0 20.0
## 4 silt 20 20.0 20.0 20.0
## 5 sand 60 60.0 60.0 60.0depth. This is the key.gather## # A tibble: 5 × 5
## property five_cm ten_cm fifteen_cm twenty_cm
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 pH 7 6.7 6.5 6.4
## 2 NO3 35 22.0 20.0 18.0
## 3 clay 20 20.0 20.0 20.0
## 4 silt 20 20.0 20.0 20.0
## 5 sand 60 60.0 60.0 60.0value. If you know what it is, name it (NO3ppm). If you have many mixed units, you can use value = value or value = measured etc.gathertidy_soil<-gather(soil, five_cm, ten_cm, fifteen_cm, twenty_cm,
key=depth, value=value)
tidy_soil## # A tibble: 20 × 3
## property depth value
## <chr> <chr> <dbl>
## 1 pH five_cm 7.0
## 2 NO3 five_cm 35.0
## 3 clay five_cm 20.0
## 4 silt five_cm 20.0
## 5 sand five_cm 60.0
## 6 pH ten_cm 6.7
## 7 NO3 ten_cm 22.0
## 8 clay ten_cm 20.0
## 9 silt ten_cm 20.0
## 10 sand ten_cm 60.0
## 11 pH fifteen_cm 6.5
## 12 NO3 fifteen_cm 20.0
## 13 clay fifteen_cm 20.0
## 14 silt fifteen_cm 20.0
## 15 sand fifteen_cm 60.0
## 16 pH twenty_cm 6.4
## 17 NO3 twenty_cm 18.0
## 18 clay twenty_cm 20.0
## 19 silt twenty_cm 20.0
## 20 sand twenty_cm 60.0filter(tidy_soil, property == "pH")%>%
ggplot(aes(x=depth, y=value))+
geom_point(size=3)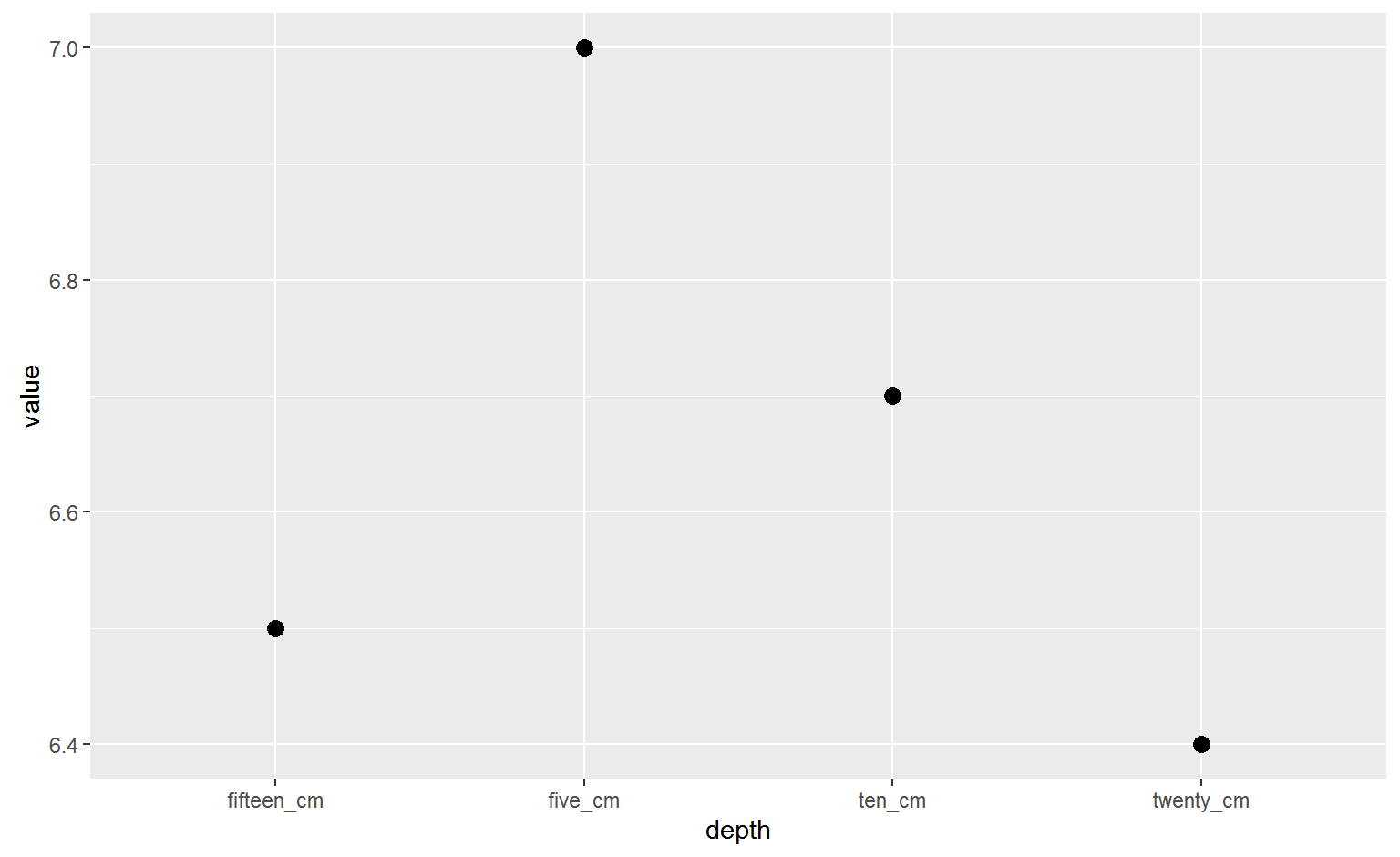 Almost!
Almost!
spreadUse this if one of your columns is full of what could be headers
## # A tibble: 6 × 4
## Year Commodity `Data Item` Value
## <int> <chr> <chr> <dbl>
## 1 2015 OATS OATS - ACRES HARVESTED 57000
## 2 2015 OATS OATS - ACRES PLANTED 125000
## 3 2015 OATS OATS - PRODUCTION, MEASURED IN $ 11027000
## 4 2015 OATS OATS - PRODUCTION, MEASURED IN BU 4161000
## 5 2015 OATS OATS - YIELD, MEASURED IN BU / ACRE 73
## 6 2014 OATS OATS - ACRES HARVESTED 55000key, here it is Data Item.value column, here they already have it as Valuesspread(oats, key = `Data Item`, value = Value)## # A tibble: 150 × 8
## Year Commodity `OATS - ACRES HARVESTED` `OATS - ACRES PLANTED`
## * <int> <chr> <dbl> <dbl>
## 1 1866 OATS 450000 NA
## 2 1867 OATS 500000 NA
## 3 1868 OATS 560000 NA
## 4 1869 OATS 610000 NA
## 5 1870 OATS 660000 NA
## 6 1871 OATS 750000 NA
## 7 1872 OATS 850000 NA
## 8 1873 OATS 910000 NA
## 9 1874 OATS 960000 NA
## 10 1875 OATS 1075000 NA
## # ... with 140 more rows, and 4 more variables: `OATS - PRODUCTION,
## # MEASURED IN $` <dbl>, `OATS - PRODUCTION, MEASURED IN BU` <dbl>, `OATS
## # - YIELD, MEASURED IN BU / ACRE` <dbl>, `OATS - YIELD, MEASURED IN BU /
## # NET PLANTED ACRE` <dbl>spread(oats, key = `Data Item`, value = Value)%>%
ggplot(aes(y=`OATS - YIELD, MEASURED IN BU / ACRE`,
x = `OATS - ACRES HARVESTED`, label = Year))+
geom_text()
## # A tibble: 4 × 7
## trt block part may june july august
## <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
## 1 0 A leaf 5.5 7.5 12.5 12.5
## 2 100 A stem 5.7 6.7 7.7 7.7
## 3 130 A fruit 0.0 2.0 8.2 8.8
## 4 150 A root 12.0 14.0 22.5 22.0Which columns need to be incorporated?
What is the key?
What is the value?
gather(biomass, may, june, july, august, key = month, value = g_m2)## # A tibble: 16 × 5
## trt block part month g_m2
## <chr> <chr> <chr> <chr> <dbl>
## 1 0 A leaf may 5.5
## 2 100 A stem may 5.7
## 3 130 A fruit may 0.0
## 4 150 A root may 12.0
## 5 0 A leaf june 7.5
## 6 100 A stem june 6.7
## 7 130 A fruit june 2.0
## 8 150 A root june 14.0
## 9 0 A leaf july 12.5
## 10 100 A stem july 7.7
## 11 130 A fruit july 8.2
## 12 150 A root july 22.5
## 13 0 A leaf august 12.5
## 14 100 A stem august 7.7
## 15 130 A fruit august 8.8
## 16 150 A root august 22.0spread(biomass_tidy, key = part, value = g_m2)## # A tibble: 16 × 7
## trt block month fruit leaf root stem
## * <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
## 1 0 A august NA 12.5 NA NA
## 2 0 A july NA 12.5 NA NA
## 3 0 A june NA 7.5 NA NA
## 4 0 A may NA 5.5 NA NA
## 5 100 A august NA NA NA 7.7
## 6 100 A july NA NA NA 7.7
## 7 100 A june NA NA NA 6.7
## 8 100 A may NA NA NA 5.7
## 9 130 A august 8.8 NA NA NA
## 10 130 A july 8.2 NA NA NA
## 11 130 A june 2.0 NA NA NA
## 12 130 A may 0.0 NA NA NA
## 13 150 A august NA NA 22.0 NA
## 14 150 A july NA NA 22.5 NA
## 15 150 A june NA NA 14.0 NA
## 16 150 A may NA NA 12.0 NA